In a recent published in Nature Microbiology, scientists have proposed a new methodology to name single-celled organisms, or prokaryotes, which uses genome sequences as types of nomenclature.
So far, regulations to name prokaryotes are defined under the International Code of Nomenclature of Prokaryotes (ICNP), but this existing system requires new species to be grown in a laboratory and at least two living or frozen samples of the microorganism to be submitted. Most prokaryotes are not available as pure cultures, and hence ineligible to be named under the ICNP regulations, the SeqCode: a nomenclatural code for prokaryotes described from sequence data research paper said.
What are prokaryotes?
Prokaryotes are the smallest form of life that can survive independently. These small organisms, mostly single-celled, lack a defined nucleus or any cell organelles due to lack of internal membranes.
Prokaryotes are divided into two domains: bacteria and archaea.
The International Code of Nomenclature of Prokaryotes and its limitations
The International Code of Nomenclature of Prokaryotes (ICNP) is an updated version of the 1990 revision of the Bacteriological Code.
It was published in the International Journal of Systematic and Evolutionary Microbiology in 2019. According to these rules, scientists must grow the species of prokaryotes in the laboratory and submit a “type” culture. According to the U.K. Health Security Agency, a or a type strain is the strain on which the description of a species is based. This requirement has “hindered the development of a nomenclature for uncultured and fastidious cultured prokaryotes”, researchers proposing the SeqCode said.
A description of the species must also be published in a scientific journal and must be accepted by the International Committee on Systematics of Prokaryotes (ICSP) that administers the ICNP.
Named prokaryotes account for less than 0.2% of the total. Excluding the uncultured majority means that a substantial portion of prokaryotes remain poorly-ordered and are often given synonymous names or alphanumeric codes.
What is SeqCode?
SeqCode, formally The Code of Nomenclature of Prokaryotes Described from Sequence Data, uses genome sequence data as a common factor for representing both cultivated and uncultivated microorganisms while rules for priority remain similar to those of the ICNP. The SeqCode also recognises the priority of names validly published under the ICNP rules provided that they do not violate the priority of names published under the new system.
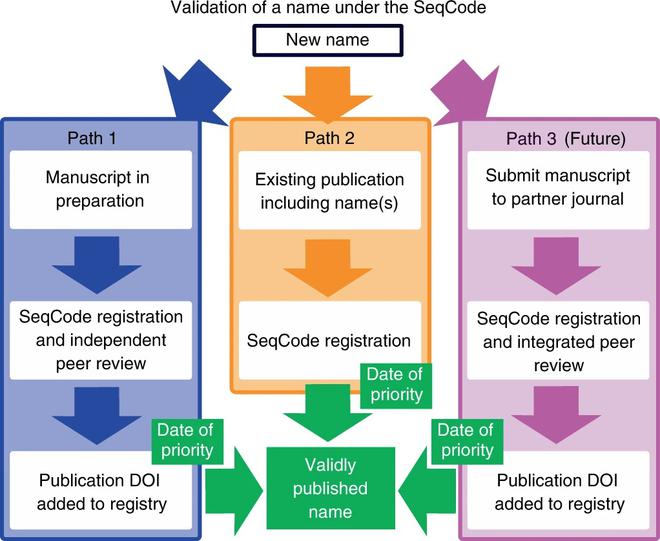
SeqCode Registry is a registration web portal that will record and validate names and nomenclatural types and link them to metadata. A draft version of the registry is already available, and all of its public data are accessible and reusable through the Creative Commons Attribution 4.0 License, except where mentioned otherwise.
There are two pathways that are currently available to register and validate names through the SeqCode Registry, but according to researchers, the best-case scenario would include entering and reviewing data before publication through a preregistration process that takes place before the initial submission or resubmission of a manuscript. This will allow SeqCode Registry to perform automated checks and provide curator inputs as well.
Why is SeqCode needed?
Scientists are of the view that ICNP’s unique restrictions regarding viable and accessible type strains have distanced quite a few microbiologists and normalised publishing names outside its regulation. SeqCode solves this problem by providing a “user-friendly resource that serves the common interests of the wider research community”, the research paper says. Authors are also of the view that SeqCode promotes “findability, accessibility, interoperability and reusability”, and its interoperable data structures will help the sharing of SeqCode names across research communities.
- Prokaryotes are the smallest form of life that can survive independently. These small organisms, mostly single-celled, lack a defined nucleus or any cell organelles due to lack of internal membranes.
- The SeqCode also recognises the priority of names validly published under the ICNP rules provided that they do not violate the priority of names published under the new system.
- Scientists are of the view that ICNP’s unique restrictions regarding viable and accessible type strains have distanced quite a few microbiologists and normalised publishing names outside its regulation.







